R and RStudio#
Users of the Esrum cluster have the option running R directly or via two RStudio servers.
Warning
The RStudio servers are only for running R. If you need to run other tasks then you must connect to the head node and run them using Slurm as described in Running jobs using Slurm.
Resource intensive tasks running on the RStudio server will likely negatively impact everyone using the service, and we may therefore terminate such tasks without warning if we deem it necessary.
R on Esrum#
This section describes steps required to use R and lays out various tips for making your work easier. See below
While it is also possible to use R on a compute node interactively, this page section focuses in particular on how to run R scripts non-interactively via Slurm in order to take full advantage of the available compute resources.
Selecting an R version#
Several versions of R are available via the module system. To load these, you need to load the version of R you want and a version of GCC, which is required to install/load R libraries.
If you intend to also make use of the RStudio servers, then we recommend
that you R/4.3.3 (or another version of R/4.3.x) with
gcc/8.5.0. This ensures that the R libraries you install are
compatible between the compute nodes and the RStudio servers.
By default, the 4.3.x versions of R loads gcc/8.5.0, so you can
simply use the --auto option when loading R/4.3.x:
$ module load --auto R/4.3.3
Loading R/4.3.3
Loading requirement: gcc/8.5.0
R modules installed using versions of R other than 4.3.x will not be
available on the RStudio server, and you will need to install them
again.
Warning
Using a GCC version greater than 8.x with R/4.3.x may cause
modules you install to fail to load on the RStudio server with the
errors similar to the following:
Error: package or namespace load failed for ‘wk’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so':
/lib64/libstdc++.so.6: version `GLIBCXX_3.4.26' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so)
See the Troubleshooting section below for more information.
Submitting R scripts using Slurm#
The recommended way to run R on Esrum is as non-interactive scripts submitted to slurm. This not only ensures that your analyses do not impact other users, but also makes your analyses reproducible.
To run an R script on the command-line, simply use the Rscript
command:
$ cat my_script.R
cat("Hello, world!\n")
$ Rscript my_script.R
Hello, world!
For simple scripts you can use the commandArgs function to pass
arguments to your scripts, allowing you to use them to process arbitrary
data-sets:
args <- commandArgs(trailingOnly = TRUE)
cat("Hello, ", args[1], "!\n", sep="")
$ Rscript my_script.R world
Hello, world!
If your script requires a heterogeneous set of input files or options to
run, then it is recommended to use an argument parser such as the
argparser R library. To use the argparser library you must first
install it using the install.packages("argparser") command.
The following shows a brief example of how you might use the
argparser library. It can also be downloaded here.
#!/usr/bin/env Rscript
library(argparser)
parser <- arg_parser("This is my script!")
parser <- add_argument(parser, "input_file", help="My data")
parser <- add_argument(parser, "--p-value", default=0.05, help="Maximum P-value")
args <- parse_args(parser)
cat("I would process the file", args$input_file, "with a max P-value of", args$p_value, "\n")
This allows you to document your command-line options, specify default values, and much more:
$ Rscript my_script.R
usage: my_script.R [--] [--help] [--opts OPTS] [--p-value P-VALUE]
input_file
This is my script!
positional arguments:
input_file My data
flags:
-h, --help show this help message and exit
optional arguments:
-x, --opts RDS file containing argument values
-p, --p-value Maximum P-value [default: 0.05]
Error in parse_args(parser) :
Missing required arguments: expecting 1 values but got 0 values: ().
Execution halted
$ Rscript my_script.R my_data.tsv
I would process the file my_data.tsv with a max P-value of 0.05
Finally, you write can write a small bash script to automatically load the required version of R and to call your script when you submit it to Slurm (using your preferred version of R):
#!/bin/bash
module load gcc/8.5.0 R/4.1.2
Rscript "${@}"
The "${@}" safely passes all your command-line arguments to
Rscript, even if they contain spaces. This wrapper script can then
be used to submit/call any of your R-scripts:
$ sbatch run_rscript.sh my_script.R my_data.tsv --p-value 0.01
Submitted batch job 18090212
$ cat slurm-18090212.out
I would process the file my_data.tsv with a max P-value of 0.01
Installing R modules#
Modules may be installed in your home folder using the
install.packages command:
$ module load gcc/8.5.0 R/4.3.1
$ R
> install.packages("ggplot2")
Warning in install.packages("ggplot2") :
'lib = "/opt/software/R/4.3.1/lib64/R/library"' is not writable
Would you like to use a personal library instead? (yes/No/cancel) yes
Would you like to create a personal library
‘/home/abc123/R/x86_64-pc-linux-gnu-library/4.3’
to install packages into? (yes/No/cancel) yes
When asked to pick a mirror, either pick 0-Cloud by entering 1
and pressing enter, or enter the number corresponding to a location near
you and press enter:
--- Please select a CRAN mirror for use in this session ---
Secure CRAN mirrors
1: 0-Cloud [https]
[...]
Selection: 1
RStudio servers#
The RStudio servers can be found at https://esrumweb01fl/rstudio/ and https://esrumweb02fl/rstudio/. You must have applied for access as described on the Applying for access page, and you must be connected via the UCPH VPN in order to connect to these servers.
If you have not been granted access, or if you are not connected via the
VPN, then you will likely see a browser error message like This site
can't be reached. See Connecting to the cluster for more information.
To log in, use the short form of your UCPH username (i.e. abc123):
RStudio server best practice#
Since the RStudio server is a shared resource where that many users may be using simultaneously, we ask that you show consideration towards other users of the server.
In particular,
Try to limit the size of the data-sets you work with on the RStudio server. Since all data has to be read from (or written to) network drives, one person reading or writing a large amount of data can cause significant slow-downs for everyone using the service.
We therefore recommend that you load a (small) subset of your data in RStudio, that you use that subset of data to develop your analyses processes, and that you use that to process your complete dataset via an R-script submitted to Slurm as described in Running jobs using Slurm.
See the R and RStudio page for additional guidance on how to use R with Slurm.
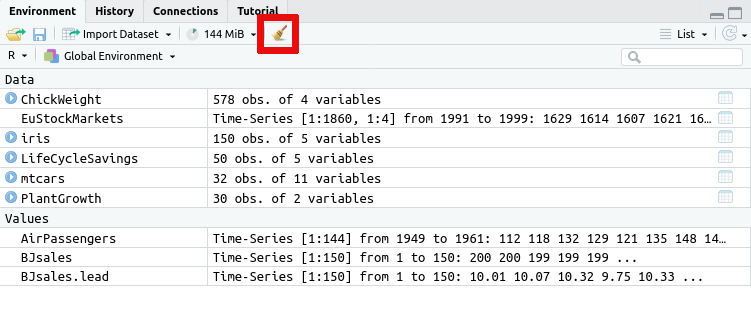
Don't keep data in memory that you do not need. Data that you no longer need can be freed with the
rmfunction or using the broom icon on theEnvironmenttab in RStudio. This also helps prevent RStudio from filling your home folder when your session is closed (see Troubleshooting below).Do not run resource intensive tasks via the embedded terminal. As noted above, such tasks will be terminated without warning if deemed to have a negative impact on other users. Instead, such tasks should be run using Slurm as described in Running jobs using Slurm.
Preserving loaded data#
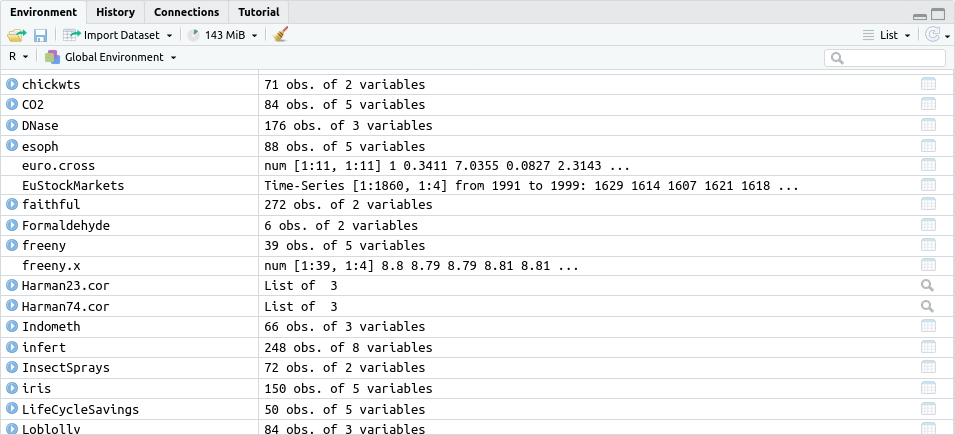
Data that you have loaded into R and other variables you have defined
are visible on the Environment tab in RStudio along with the amount
of memory used (here 143 MiB):
By default, this data will be saved to your RStudio folder on the
/scratch drive when you quit your session or when it automatically
suspends after 9 hours of inactivity. This may, however, result in very
large amounts of data being saved to disk and, consequently, large of
amounts of data having to be read when you log in again, resulting in
logging in taking a very long time.
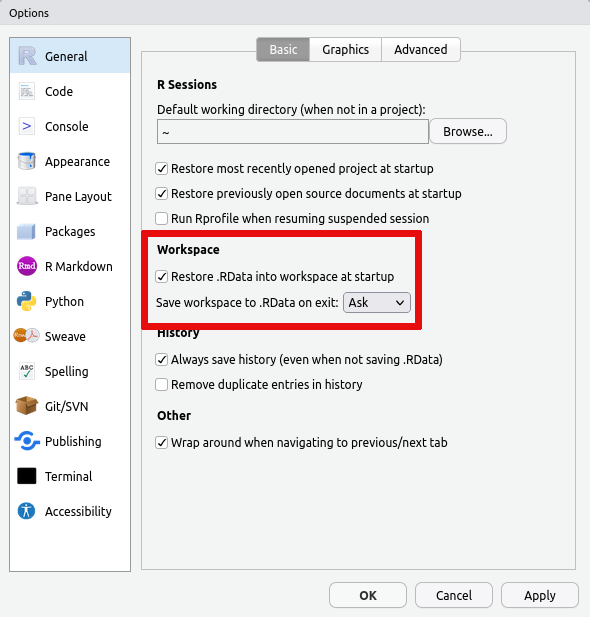
For this reason we recommend disabling the saving and loading of
.RData in the Global Settings accessible via the Tools Menu
as shown:
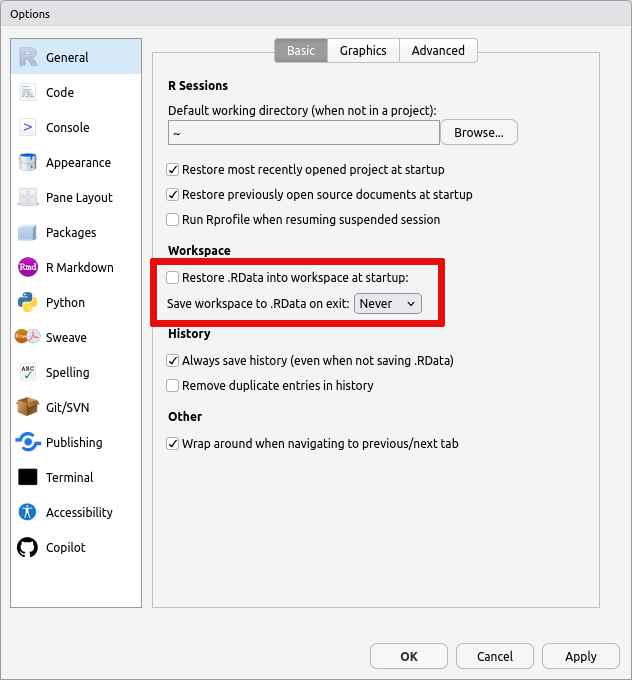
This ensures that you always start with a fresh session and that you therefore are able to log in quickly to the RStudio server.
It is also recommended that the Always save history (even when not
saving .RData) option is enabled, as the commands you type into the R
terminal will otherwise not be saved.
Use of GitHub Copilot#
You can make use of GitHub Copilot in RStudio, provided that you have a valid license. Please refer to the guidelines on use of generative AI and LLMs before doing so.
To enable Copilot, click on the
Toolsmenu and selectGlobal Options: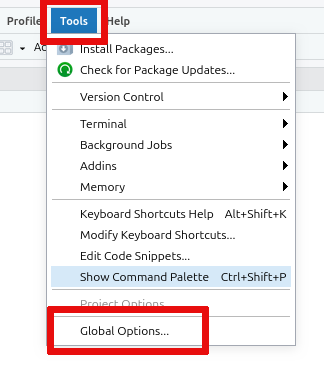
Open the
Copilottab and tick theEnable Github Copilotcheckbox. Then click theSign Inbutton: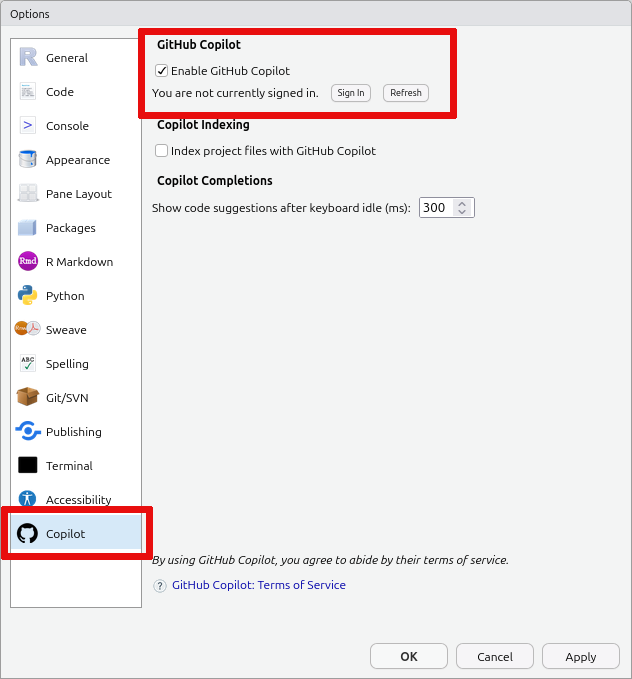
Finally, copy the verification code you are shown, click on the displayed link, and follow the instructions on Github.
Troubleshooting#
libstdc++.so.6: version 'GLIBCXX_3.4.26' not found#
If you build an R library on the head/compute nodes using a version of
the GCC module other than gcc/8.5.0, then this library may fail to
load on the RStudio node or when gcc/8.5.0 is loaded on the
head/compute nodes:
$ R
> library(wk)
Error: package or namespace load failed for ‘wk’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so':
/lib64/libstdc++.so.6: version `GLIBCXX_3.4.26' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so)
To fix his, you will need to reinstall the affected R libraries using one of two methods:
Connect to the RStudio server as described in the RStudio servers section, and simply install the affected packages using the
install.packagesfunction:> install.packages("wk")You may need to repeat this step multiple times, for every package that fails to load.
Connect to the head node or a compute node, and take care to load the correct version of GCC before loading R:
$ module load gcc/8.5.0 R/4.3.2 $ R > install.packages("wk")
The name of the affected module can be determined by looking at the
error message above. In particular, the path
/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so
contains a pair of folders named R/x86_64-pc-linux-gnu-library,
which specifies the kind of system we are running on. Immediately after
that we find the package name, namely wk in this case.
You can identify all affected packages in your "global" R library by running the following commands:
$ module load gcc/8.5.0 R/4.3.2
cdto your R library$ cd ~/R/x86_64-pc-linux-gnu-library/4.3/
Test every installed library
$ for lib in $(ls);do echo "Testing ${lib}"; Rscript <(echo "library(${lib})") > /dev/null;done
Output will look like the following:
Testing httpuv
Testing igraph
Error: package or namespace load failed for ‘igraph’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/igraph/libs/igraph.so':
/opt/software/gcc/8.5.0/lib64/libstdc++.so.6: version `GLIBCXX_3.4.29' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/igraph/libs/igraph.so)
Execution halted
Testing isoband
Error: package or namespace load failed for ‘isoband’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/isoband/libs/isoband.so':
/opt/software/gcc/8.5.0/lib64/libstdc++.so.6: version`GLIBCXX_3.4.29' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/isoband/libs/isoband.so)
Execution halted
Testing labeling
Testing later
Locate the error messages like the one shown above in the output and
reinstall the affected libraries using the install.packages command:
$ R
> install.packages(c("igraph", "isoband"))
Incorrect or invalid username/password#
Please make sure that you are entering your username in the short form
(i.e. abc123) and that you have applied for and been given access to
the Esrum HPC (see Applying for access). If the problem
persists, please Contact us us for assistance.
Logging in takes a very long time#
Similar to regular R, RStudio will automatically save the data you have loaded into your R session and will restore it when you return later, so that you can continue your work. However, this many result in large amounts of data being saved and loading this data may result in a large delay when you attempt to log in at a later date.
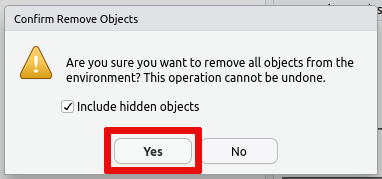
It is therefore recommended that you regularly clean up your workspace using the built-in tools, when you no longer need to have the data loaded in R.
You can remove individual bits of data using the rm function in R.
This works both when using regular R and when using RStudio. The
following gives two examples of using the rm function, one removing
a single variable and the other removing all variables in the current
session:
# 1. Remove the variable `my_variable`
rm(my_variable)
# 2. Remove all variables from your R session
rm(list = ls())
Alternatively you can remove all data saved in your R session using the
broom icon on the Environment tab:
If you wish to prevent this issue in the first case, then you can also
turn off saving the data in your session on exit and/or turn off loading
the saved data on startup. This is accomplished via the Global
Options... accessible from the Tools menu:
Should your R session have grown to such a size that you simply cannot log in and clean it up, then it may be necessary to remove the files containing the data that R/RStudio has saved. This data is stored in two locations:
In the
.RDatafile in your home (~/.RData). This is where R saves your data if you answer yesSave workspace image? [y/n/c]when quitting R.In the
environmentfile in your RStudio session folder (~/.local/share/rstudio/sessions/active/session-*/suspended-session-data/environment). This is where RStudio saves your data should your login time out while using RStudio.
Please Contact us us if you need help removing the correct files.
libstdc++.so.6: version 'GLIBCXX_3.4.26' not found#
See the troubleshooting section on the R and RStudio page.