Troubleshooting¶
This page gathers all troubleshooting steps from the other parts of this documentation for easy access. Remember that you are always welcome to contact us if you have questions or problems relating to the cluster.
Connecting to Esrum¶
If you have not already been granted access to the server, then please see the Applying for access page before continuing!
Timeout while connecting to the cluster¶
You may experience timeout errors when you attempt to connect to Esrum.
On Linux, this typically results in an Operation timed out message:
$ ssh abc123@esrumhead01fl.unicph.domain
ssh: connect to host esrumhead01fl.unicph.domain port 22: Operation timed out
On Windows, using MobaXterm, it may result in a connection timed out
message:
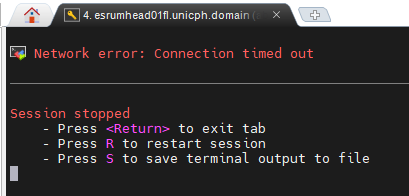
Firstly verify that you are correctly connected to the UCPH VPN. This is required to connect to Esrum. See the Connecting to the cluster page for more information.
If you are still unable to connect to Esrum after verifying that you are correctly connected to the UCPH network, then please try to visit either our Project Manager or our Cohort Catalog.
If you are able to visit either of the Project Manager or Cohort Catalog pages, then you most likely do not have proper permissions to connect to Esrum. Please contact us and we will provide further guidance.
If you are unable to connect to the VPN or to either of the above pages
while connected to the VPN, then there may be other problems with your
account. We recommend that you either contact us for
assistance or, if you prefer, that you submit a ticket to the UCPH-IT
Serviceportal, using the Research Applications Counseling and
Support / Forskningsapplikationer Rådgivning og support ticket
category.
File uploads using MobaXterm never start¶
Please make sure that your session is configured to use the SCP
(enhanced speed) browser type. See step 4 in the
Configuring MobaXterm section.
Network-folders in ~/ucph are not available¶
Please make sure that you have disabled use of GSSAPI Kerberos as
described in the Configuring MobaXterm section. Similarly, if
using Linux or OSX, then you cannot be authenticating using a Kerberos
ticket.
OpenConnect fails to perform two-factor authentication¶
OpenConnect may fail to connect to the KU VPN when using the default Configuration. In that case, when connecting, OpenConnect will make several connection attempts, allow you to enter your password, and then fail before you are able to use your two-factor authentication method:
POST https://vpn.ku.dk/
Connected to 130.225.226.54:443
SSL negotiation with vpn.ku.dk
Connected to HTTPS on vpn.ku.dk with ciphersuite (TLS1.3)-(ECDHE-SECP256R1)-(RSA-PSS-RSAE-SHA256)-(AES-128-GCM)
Got HTTP response: HTTP/1.1 404 Not Found
Unexpected 404 result from server
GET https://vpn.ku.dk/
Connected to 130.225.226.54:443
SSL negotiation with vpn.ku.dk
Connected to HTTPS on vpn.ku.dk with ciphersuite (TLS1.3)-(ECDHE-SECP256R1)-(RSA-PSS-RSAE-SHA256)-(AES-128-GCM)
Got HTTP response: HTTP/1.0 302 Temporary moved
GET https://vpn.ku.dk/+webvpn+/index.html
SSL negotiation with vpn.ku.dk
Connected to HTTPS on vpn.ku.dk with ciphersuite (TLS1.3)-(ECDHE-SECP256R1)-(RSA-PSS-RSAE-SHA256)-(AES-128-GCM)
Please enter your username and password.
Password: ************
POST https://vpn.ku.dk/+webvpn+/index.html
Please enter the TOTP code generated on your device
POST https://vpn.ku.dk/+webvpn+/login/challenge.html
Please enter the TOTP code generated on your device
POST https://vpn.ku.dk/+webvpn+/login/challenge.html
Please enter the TOTP code generated on your device
3 consecutive empty forms, aborting loop
Failed to complete authentication
In this happens, then make sure that you are using the
--useragent=AnyConnect --no-external-auth options as shown in the
Connecting on Linux section.
Slurm basics¶
Error: Unable to allocate resources: Invalid account or account/partition combination specified¶
If you get this error while using sbatch or srun, then ensure
that your sbatch or srun, then please ensure that you are not
manually specifying an account.
If you are not specifying an account, but still get this error, then please contact us. Normally, a Slurm account should automatically be created for you, but in some cases that may not have happened, and we may have to fix it manually.
Error: Requested node configuration is not available¶
If you request too many CPUs (more than 128), or too much RAM (more than 1993 GB for compute nodes and more than 3920 GB for the GPU node), then Slurm will report that the request cannot be satisfied.
If more than 128 CPUs requested:
$ sbatch --cpus-per-task 200 my_script.sh
sbatch: error: CPU count per node can not be satisfied
sbatch: error: Batch job submission failed: Requested node configuration is not available
More than 1993 GB RAM requested on compute node:
$ sbatch --mem 2000G my_script.sh
sbatch: error: Memory specification can not be satisfied
sbatch: error: Batch job submission failed: Requested node configuration is not available
To solve this, simply reduce the number of CPUs and/or the amount of RAM requested to fit within the limits described above. If your task does require more than 1993 GB of RAM, then you need to run your task on the GPU queue as described on the GPU / high-memory jobs page.
Additionally, you may receive this message if you request GPUs without specifying the correct queue or if you request too many GPUs.
If --partition=gpuqueue not specified:
$ srun --gres=gpu:2 -- echo "Hello world!"
srun: error: Unable to allocate resources: Requested node configuration is not available
If more than 2 GPUs requested:
$ srun --partition=gpuqueue --gres=gpu:3 -- echo "Hello world!"
srun: error: Unable to allocate resources: Requested node configuration is not available
To solve this error, simply avoid requesting more than 2 GPUs, and
remember to include the --partition option. See also the
GPU / high-memory jobs section.
srun fails with /bin/slurm_bcast_123456.0_esrumcmpn01fl: No such file or directory¶
If you accidentally specify a folder as the first component of an
srun command, then Slurm will fail with an error message complaining
that a slurm_bcast_* executable in that folder could not be found,
where the executable name contains the job ID and the node on which it
was run:
$ srun --pty /bin/
slurmstepd: error: execve(): /bin/slurm_bcast_123456.0_esrumcmpn01fl: No such file or directory
srun: error: esrumcmpn01fl: task 0: Exited with exit code 2
To fix this, ensure that you are running an executable and not a folder:
$ srun --pty /bin/bash
Note
This failure relates to the --bcast option, that allow you to
copy an executable from the head node to a folder on the node on
which the job is executed. This is typically not required on Esrum,
since all home, project, and dataset folders are shared across nodes.
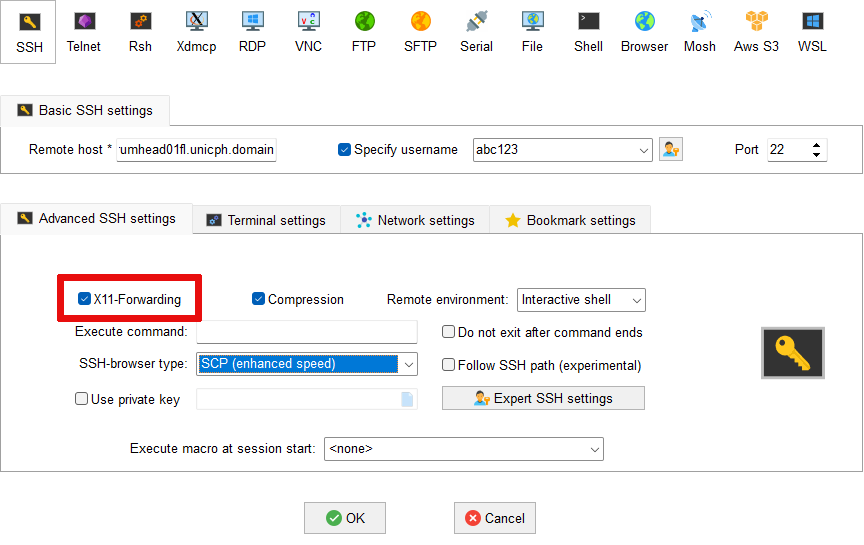
X11 forwarding not working in MobaXterm¶
Firstly right-click on Esrum in the list of User sessions and
select Edit session. Make sure that the Advanced SSH settings
tab is open and verify that X11 forwarding is enabled as shown:
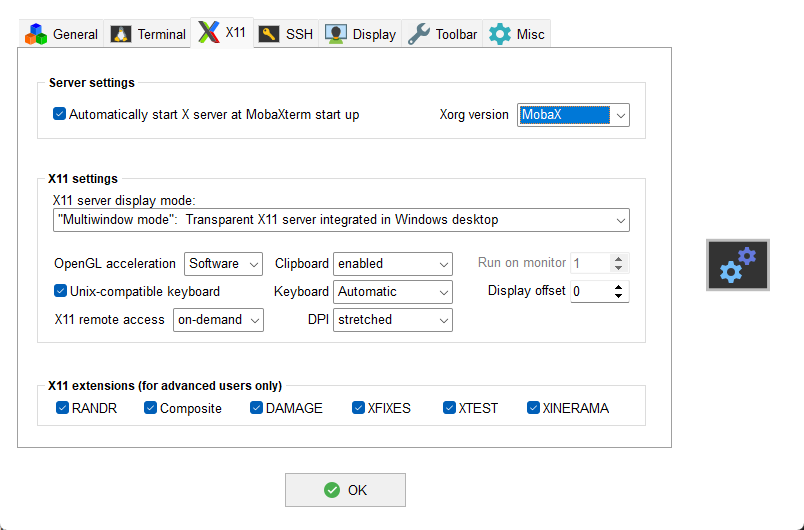
Secondly, press the OK button and open the Settings via the
gears icon on the main toolbar. Then select the X11 tab and verify
that X11 support is configured as shown:
Monitoring slurm jobs¶
Connection refused when running sacct / sacct-usage¶
If you attempt to run sacct or sacct-usage on any other node
than the head node, then you may get an error message like this:
sacct: error: slurm_persist_conn_open_without_init: failed to open persistent connection to host:localhost:6819: Connection refused
sacct: error: Sending PersistInit msg: Connection refused
sacct: error: Problem talking to the database: Connection refused
To avoid this, only run sacct and sacct-usage on the head node.
Internal data transfers on Esrum¶
rsync fails with Permission denied when copying from /datasets¶
If you forget to use the --no-perms option when rsync'ing data out
of a /datasets folder, then all permissions will be set to 000.
In other words, nobody can read, write, or execute those files and
folders.
To fix this, first run the following commands to fix the permissions,
where /path/to/copied/data is the path to the copy of the data that
you have created.
chmod -R +rX,u+w /path/to/copied/data
This will recursively mark files and folders readable for everyone, mark folders executable for everyone (required to browse them), and mark files and folders writable for you (and only you).
Then re-run rsync and remember to include the --no-perms option.
Permission denied when accessing data copied from /datasets¶
See above.
External data transfers on Esrum¶
Chains not found when connecting to sftp.ku.dk¶
If you try and fail to log in to sftp.ku.dk too many times in a short period of time, then your account may get locked out. In that case, please create a ticket using KU-IT serviceportal.
Network drives¶
The ~/ucph folder or subfolders are missing¶
Note that the ~/ucph folder is only available on the head node
(esrumhead01fl), and not on the RStudio servers nor on the compute
nodes. See the Accessing network drives from compute nodes section for how to
access the drives elsewhere.
If you are connected to the head node, then firstly make sure that you are not using GSSAPI (Kerberos) to log in. See the Connecting to the cluster page for instructions for how to disable this feature if you are using MobaXterm.
Once you have logged in to Esrum without GSSAPI enabled, and if the folder(s) are still missing, then run the following command to create any missing network folders:
$ bash /etc/profile.d/symlink-ucphmaps.sh
Once this is done, you should have a ucph symlink in your home
folder containing links to hdir (H:), ndir (N:), and
sdir (S:).
No such file or directory when accessing network drives¶
If you get a No such file or directory error when attempting to
access the network drives (~/ucph/hdir, ~/ucph/ndir, or
~/ucph/sdir), then please make sure that you are not logging in
using Kerberos (GSSAPI). See the Accessing network drives via MobaXterm
section for instructions for how to disable this feature if you are
using MobaXterm.
Note also that your login is also valid for about 10 hours, after which you will lose access to the network drives. See the section (Re)activating access to the network drives for how to re-authenticate if your access has timed out.
kinit: Unknown credential cache type while getting default ccache¶
The kinit command may fail if you are using a conda environment:
(base) $ kinit
kinit: Unknown credential cache type while getting default ccache
To circumvent this problem, either specify the full path to the
kinit executable (i.e. /usr/bin/kinit) or deactivate the
current/base environment by running conda deactivate until conda is
completely deactivated.
R¶
libstdc++.so.6: version 'GLIBCXX_3.4.26' not found¶
If you build an R library on the head/compute nodes using a version of
the GCC module other than gcc/8.5.0, then this library may fail to
load on the RStudio node or when gcc/8.5.0 is loaded on the
head/compute nodes:
$ R
> library(wk)
Error: package or namespace load failed for ‘wk’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so':
/lib64/libstdc++.so.6: version `GLIBCXX_3.4.26' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so)
To fix his, you will need to reinstall the affected R libraries using one of two methods:
Connect to the RStudio server as described in the Troubleshooting section, and simply install the affected packages using the
install.packagesfunction:> install.packages("wk")You may need to repeat this step multiple times, for every package that fails to load.
Connect to the head node or a compute node, and take care to load the correct version of GCC before loading R:
$ module load gcc/8.5.0 R/4.3.2 $ R > install.packages("wk")
The name of the affected module can be determined by looking at the
error message above. In particular, the path
/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/wk/libs/wk.so
contains a pair of folders named R/x86_64-pc-linux-gnu-library,
which specifies the kind of system we are running on. Immediately after
that we find the package name, namely wk in this case.
You can identify all affected packages in your "global" R library by running the following commands:
$ module load gcc/8.5.0 R/4.3.2
cdto your R library$ cd ~/R/x86_64-pc-linux-gnu-library/4.3/
Test every installed library
$ for lib in $(ls);do echo "Testing ${lib}"; Rscript <(echo "library(${lib})") > /dev/null;done
Output will look like the following:
Testing httpuv
Testing igraph
Error: package or namespace load failed for ‘igraph’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/igraph/libs/igraph.so':
/opt/software/gcc/8.5.0/lib64/libstdc++.so.6: version `GLIBCXX_3.4.29' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/igraph/libs/igraph.so)
Execution halted
Testing isoband
Error: package or namespace load failed for ‘isoband’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/abc123/R/x86_64-pc-linux-gnu-library/4.3/isoband/libs/isoband.so':
/opt/software/gcc/8.5.0/lib64/libstdc++.so.6: version`GLIBCXX_3.4.29' not found (required by /home/abc123/R/x86_64-pc-linux-gnu-library/4.3/isoband/libs/isoband.so)
Execution halted
Testing labeling
Testing later
Locate the error messages like the one shown above in the output and
reinstall the affected libraries using the install.packages command:
$ R
> install.packages(c("igraph", "isoband"))
Rstudio¶
"Error: Unexpected response from server" when uploading files¶
Using RStudio to upload files that are 50 MB or larger may fail with the
error message Unexpected response from server. Instead, use the
methods described on the Transferring data page to transfer files to
Esrum.
Incorrect or invalid username/password¶
Please make sure that you are entering your username in the short form
(i.e. abc123) and that you have applied for and been given access to
the Esrum HPC (see Applying for access). If the problem
persists, please contact us for assistance.
Logging in takes a very long time¶
Similar to regular R, RStudio will automatically save the data you have loaded into your R session and will restore it when you return later, so that you can continue your work. However, this many result in large amounts of data being saved and loading this data may result in a large delay when you attempt to log in at a later date.
It is therefore recommended that you regularly clean up your workspace using the built-in tools, when you no longer need to have the data loaded in R.
You can remove individual bits of data using the rm function in R.
This works both when using regular R and when using RStudio. The
following gives two examples of using the rm function, one removing
a single variable and the other removing all variables in the current
session:
1# 1. Remove the variable `my_variable`
2rm(my_variable)
3
4# 2. Remove all variables from your R session
5rm(list = ls())
Alternatively you can remove all data saved in your R session using the
broom icon on the Environment tab:
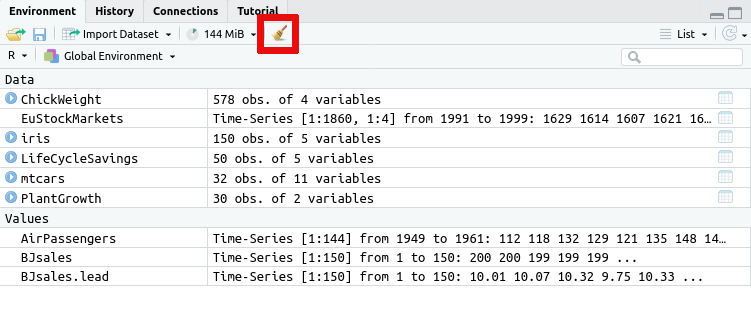
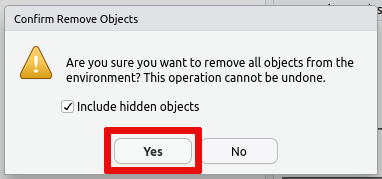
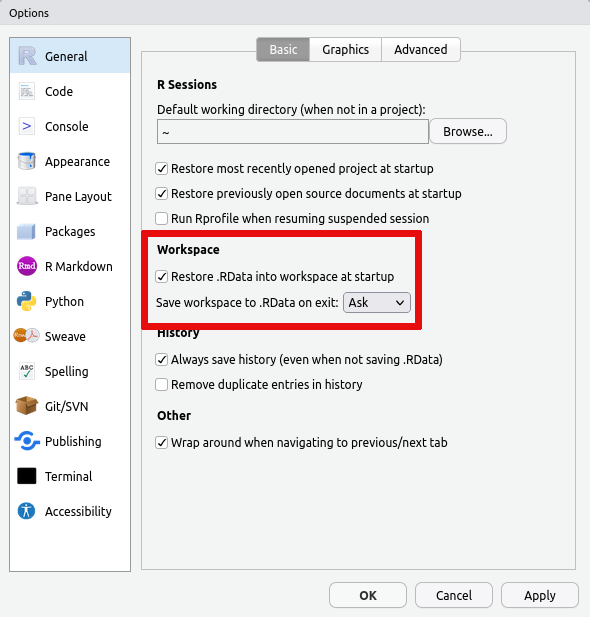
If you wish to prevent this issue in the first case, then you can also
turn off saving the data in your session on exit and/or turn off loading
the saved data on startup. This is accomplished via the Global
Options... accessible from the Tools menu:
Should your R session have grown to such a size that you simply cannot log in and clean it up, then it may be necessary to remove the files containing the data that R/RStudio has saved. This data is stored in two locations:
In the
.RDatafile in your home (~/.RData). This is where R saves your data if you answer yesSave workspace image? [y/n/c]when quitting R.In the
environmentfile in your RStudio session folder (~/.local/share/rstudio/sessions/active/session-*/suspended-session-data/environment). This is where RStudio saves your data should your login time out while using RStudio.
Please contact us us if you need help removing the correct files.
libstdc++.so.6: version 'GLIBCXX_3.4.26' not found¶
See the troubleshooting section on the Using R on Esrum page.
Jupyter Notebooks¶
Jupyter Notebooks: Browser error when opening URL¶
Depending on your browser you may receive one of the following errors. The typical causes are listed, but the exact error message will depend on your browser. It is therefore helpful to review all possible causes listed here.
When using Chrome, the cause is typically listed below the line that says "This site can't be reached".
The connection was resetThis typically indicates that Jupyter Notebook isn't running on the server, or that it is running on a different port than the one you've forwarded. Check that Jupyter Notebook is running and make sure that your forwarded ports match those used by Jupyter Notebook on Esrum.
Localhost refused to connectorUnable to connectThis typically indicates that port forwarding isn't active, or that you have entered the wrong port number in your browser. Therefore,
Verify that port forwarding is active: On OSX/Linux that means verifying that an
sshcommand is running as described in the Port forwarding for OSX/Linux users section, and on Windows that means activating port forwarding in MobaXterm as described in the Port forwarding for Windows users section.If using the instructions for Linux/OSX, verify that you ran the
sshcommand on your laptop or desktop, and not on the Esrum head node.Verify that either of these are using the same port number as in the
jupytercommand you ran or as in thehttp://127.0.0.1URL printed by Jupyter.Verify that you are using the second URL that Jupyter prints on the terminal, namely the URL starting with
http://127.0.0.1:XXXX:To access the notebook, open this file in a browser: file:///home/abc123/.local/share/jupyter/runtime/nbserver-2082873-open.html Or copy and paste one of these URLs: http://esrumcmpn07fl.unicph.domain:XXXXX/?token=0123456789abcdefghijklmnopqrstuvwxyz or http://127.0.0.1:XXXXX/?token=0123456789abcdefghijklmnopqrstuvwxyzFor security reasons it is not possible to connect directly to the compute nodes.
Snakemake¶
sacct: error: Problem talking to the database: Connection refused¶
If you are running Snakemake with the --slurm option on a compute
node, i.e. not the head node, then you will receive errors such as the
following:
Job 0 has been submitted with SLURM jobid 512921 (log: .snakemake/slurm_logs/rule_foo/512921.log).
The job status query failed with command: sacct -X --parsable2 --noheader --format=JobIdRaw,State --name 2d898259-73e4-435d-aa77-44dc44d84c1b
Error message: sacct: error: slurm_persist_conn_open_without_init: failed to open persistent connection to host:localhost:6819: Connection refused
sacct: error: Sending PersistInit msg: Connection refused
sacct: error: Problem talking to the database: Connection refused
To solve this, simply start your Snakemake pipeline on the head node
when using the --slurm option.